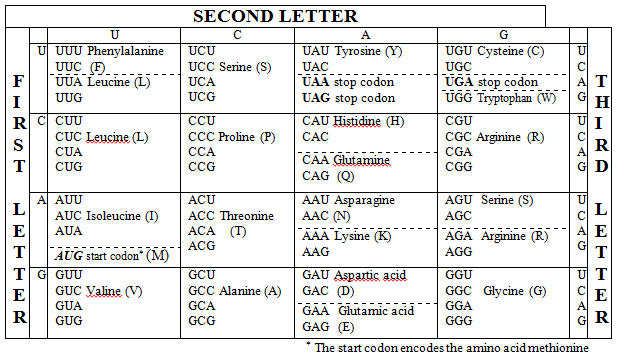
The Genetic Code and Its Universality
The genetic code is the set of rules cells use to translate nucleotide sequences in mRNA into amino acid sequences in proteins. It's nearly universal across all domains of life, which points strongly to a common evolutionary origin for all organisms on Earth.
How the Code Works
The code is read in codons, groups of three nucleotides on the mRNA. Each codon specifies either a particular amino acid or a stop signal. With four possible nucleotides at each of three positions, there are possible codons. Since only 20 amino acids need to be encoded (plus stop signals), the code is degenerate, meaning multiple codons can specify the same amino acid.
Universality and Exceptions
Most organisms, from bacteria to humans, read the genetic code the same way. That said, small deviations exist. Mitochondria and certain protozoans reassign a few codons to different amino acids or stop signals. These are the exception, not the rule.

Protein Synthesis (Translation) Process
Translation is the process of reading an mRNA sequence and building a polypeptide chain from it. It occurs in three stages: initiation, elongation, and termination.
Key Steps in Protein Synthesis
1. Initiation
- The small ribosomal subunit binds to the mRNA near the start codon (AUG), assisted by initiation factors.
- The initiator tRNA, carrying methionine (or formylmethionine in prokaryotes), base-pairs with the start codon.
- The large ribosomal subunit joins, forming the complete ribosome with the initiator tRNA seated in the P site.
2. Elongation
Before elongation begins, aminoacyl-tRNA synthetases charge each tRNA with its correct amino acid. This "charging" step is critical for accuracy because the synthetase must match the right amino acid to the right tRNA.
- A charged aminoacyl-tRNA enters the A site, where its anticodon base-pairs with the mRNA codon.
- A peptide bond forms between the amino acid in the A site and the growing polypeptide chain held in the P site. The ribosome catalyzes this reaction.
- The ribosome translocates one codon forward. The tRNA that was in the A site moves to the P site (now carrying the polypeptide), and the empty tRNA in the P site shifts to the E site and exits.
- This cycle repeats, with GTP hydrolysis providing energy for tRNA binding and translocation.
3. Termination
- The ribosome encounters a stop codon (UAA, UAG, or UGA). No tRNA recognizes these codons.
- Release factors bind to the A site instead, triggering hydrolysis of the bond between the polypeptide and the final tRNA.
- The completed polypeptide is released, and the ribosomal subunits dissociate from the mRNA.
Ribosome Structure and Function
Ribosomes have three internal sites that tRNAs move through during elongation:
- A site (aminoacyl-tRNA site): Where the incoming charged tRNA first binds. The tRNA anticodon must correctly base-pair with the mRNA codon here, ensuring the right amino acid is added.
- P site (peptidyl-tRNA site): Holds the tRNA attached to the growing polypeptide chain.
- E site (exit site): Holds the now-empty (deacylated) tRNA just before it leaves the ribosome.
Think of it as a conveyor belt: tRNAs enter at A, do their work at P, and leave at E.
Prokaryotic vs. Eukaryotic Translation
While the core mechanism is the same, there are several clinically and biologically important differences:
| Feature | Prokaryotes | Eukaryotes |
|---|---|---|
| Ribosome size | 70S (30S + 50S subunits) | 80S (40S + 60S subunits) |
| mRNA recognition | Shine-Dalgarno sequence upstream of start codon recruits the ribosome | 5' cap and poly-A tail guide ribosome binding |
| Coupling | Transcription and translation happen simultaneously in the cytoplasm | Transcription occurs in the nucleus; mRNA is exported to the cytoplasm for translation |
| Initiator amino acid | Formylmethionine (fMet) | Methionine (Met) |
| Antibiotic targeting | Many antibiotics (e.g., streptomycin, chloramphenicol, tetracycline) specifically target 70S ribosomes | 80S ribosomes are not affected by these antibiotics, which is why they can treat bacterial infections without harming human cells |
The difference in ribosome structure is especially important in medicine. Because prokaryotic and eukaryotic ribosomes differ in size and shape, antibiotics can selectively inhibit bacterial translation while leaving human ribosomes unharmed. This is the basis of selective toxicity for many commonly used antibiotics.