Dynamic programming is a powerful technique in bioinformatics, breaking complex problems into simpler subproblems. It's crucial for efficient algorithms in sequence analysis, enabling faster and more accurate results in tasks like alignment and structure prediction.
Key principles include optimal substructure, recursive definition, and memoization. These concepts form the foundation for solving various bioinformatics problems, from sequence alignment to RNA folding, by efficiently reusing solutions to smaller problem instances.
Fundamentals of dynamic programming
- Dynamic programming optimizes complex problems by breaking them into simpler subproblems, crucial for efficient bioinformatics algorithms
- Applies to various sequence analysis tasks in bioinformatics, enabling faster and more accurate results
- Utilizes recursive problem-solving techniques to build solutions incrementally, reducing computational complexity
Key principles
- Optimal substructure allows breaking down problems into smaller, solvable components
- Recursive definition forms the basis for dynamic programming solutions
- Memoization stores previously computed results to avoid redundant calculations
- Bottom-up approach builds solutions from smallest subproblems to larger ones
- Principle of optimality ensures global optimum through local optimal choices
Optimal substructure
- Defines problems where optimal solution contains optimal solutions to subproblems
- Enables efficient problem-solving by reusing solutions to smaller instances
- Crucial for dynamic programming algorithms in sequence alignment and structure prediction
- Allows for recursive formulation of problems, leading to efficient solutions
- Applies to various bioinformatics problems (sequence alignment, RNA folding)
Overlapping subproblems
- Occurs when a recursive algorithm revisits the same subproblems repeatedly
- Enables memoization to store and reuse solutions, reducing computational complexity
- Common in sequence comparison algorithms (local and global alignment)
- Facilitates efficient solutions for problems with exponential naive recursive approaches
- Identifies opportunities for optimization in bioinformatics algorithms
Applications in bioinformatics
- Dynamic programming forms the foundation for numerous critical bioinformatics algorithms and tools
- Enables efficient analysis of biological sequences, structures, and relationships between molecules
- Facilitates discovery of evolutionary relationships and functional predictions in genomics and proteomics
Sequence alignment
- Compares DNA, RNA, or protein sequences to identify similarities and differences
- Global alignment aligns entire sequences (Needleman-Wunsch algorithm)
- Local alignment finds regions of high similarity within sequences (Smith-Waterman algorithm)
- Pairwise alignment compares two sequences, while multiple sequence alignment compares three or more
- Applications include:
- Identifying homologous genes or proteins
- Detecting evolutionary relationships between organisms
- Predicting protein structure and function
RNA structure prediction
- Predicts secondary structure of RNA molecules based on their primary sequence
- Utilizes dynamic programming to find optimal base-pairing configurations
- Nussinov algorithm predicts maximum number of base pairs in RNA structure
- Zuker algorithm incorporates energy minimization for more accurate predictions
- Applications include:
- Studying RNA folding and stability
- Identifying functional RNA elements (riboswitches, regulatory regions)
- Designing RNA-based therapeutics
Gene finding
- Identifies coding regions within genomic DNA sequences
- Uses dynamic programming to evaluate potential exon-intron structures
- Hidden Markov Models (HMMs) often employed for gene prediction
- Incorporates various signals (start codons, splice sites, stop codons) in prediction process
- Applications include:
- Annotating newly sequenced genomes
- Identifying novel genes and their structures
- Comparative genomics studies
Dynamic programming algorithms
- Form the core of many bioinformatics analysis tools and techniques
- Enable efficient solutions to complex sequence analysis problems
- Provide foundation for more advanced algorithms and machine learning approaches in bioinformatics
Needleman-Wunsch algorithm
- Global sequence alignment algorithm for pairwise comparison
- Constructs a scoring matrix to find optimal alignment of entire sequences
- Uses dynamic programming to fill the matrix and traceback for alignment
- Time complexity for sequences of length m and n
- Applications include:
- Comparing protein or DNA sequences from different species
- Identifying conserved regions across entire gene or protein sequences
Smith-Waterman algorithm
- Local sequence alignment algorithm for finding regions of high similarity
- Modifies Needleman-Wunsch by allowing alignment to start and end anywhere in sequences
- Introduces zero as minimum score to identify local alignments
- More sensitive than heuristic methods (BLAST) but computationally intensive
- Applications include:
- Identifying conserved domains or motifs within proteins
- Detecting partial matches in database searches
Nussinov algorithm
- Predicts RNA secondary structure by maximizing base pair count
- Uses dynamic programming to fill a matrix of optimal substructures
- Time complexity for RNA sequence of length n
- Serves as foundation for more sophisticated RNA structure prediction algorithms
- Applications include:
- Initial step in more complex RNA folding algorithms
- Studying basic principles of RNA structure formation

Scoring matrices
- Essential components of sequence alignment algorithms in bioinformatics
- Quantify the similarity or dissimilarity between biological sequence elements
- Guide alignment algorithms in making optimal choices during sequence comparison
PAM vs BLOSUM
- PAM (Point Accepted Mutation) matrices based on evolutionary models
- Derived from closely related protein sequences
- Higher PAM numbers indicate greater evolutionary distance
- BLOSUM (BLOcks SUbstitution Matrix) matrices based on observed substitutions
- Derived from more distantly related sequences
- Lower BLOSUM numbers indicate more conserved alignments
- Choice between PAM and BLOSUM depends on expected evolutionary distance between sequences
- PAM matrices often used for closely related sequences, BLOSUM for more divergent ones
Gap penalties
- Penalize insertions or deletions (indels) in sequence alignments
- Linear gap penalties assign fixed cost for each gap position
- Affine gap penalties use different costs for gap opening and extension
- More biologically realistic, as indels often occur in blocks
- Gap penalty values affect alignment sensitivity and specificity
- Proper tuning of gap penalties crucial for accurate sequence alignment results
Substitution scores
- Quantify likelihood of one amino acid or nucleotide substituting for another
- Based on observed frequencies of substitutions in known homologous sequences
- Positive scores indicate favorable substitutions, negative scores unfavorable ones
- Incorporate biological and chemical properties of amino acids or nucleotides
- Examples include:
- BLOSUM62 matrix commonly used for protein sequence alignment
- Transition/transversion bias in DNA substitution models
Implementation techniques
- Critical for developing efficient and scalable bioinformatics algorithms
- Enable processing of large-scale genomic and proteomic datasets
- Balance between time efficiency and memory usage in algorithm design
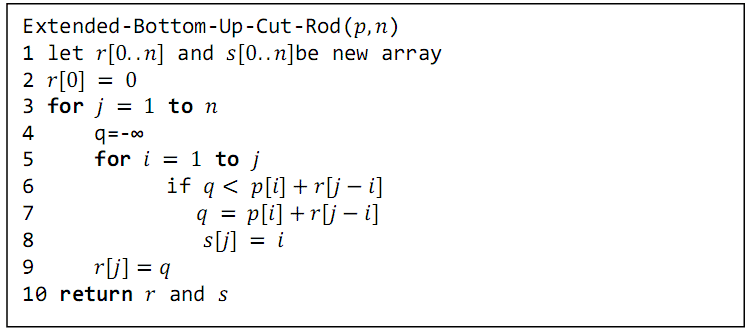
Tabulation vs memoization
- Tabulation (bottom-up approach)
- Iteratively fills a table with solutions to subproblems
- Guarantees all subproblems are solved before they are needed
- Often more efficient in terms of constant factors
- Memoization (top-down approach)
- Recursively solves problems and stores results for reuse
- Only computes subproblems that are actually needed
- Can be more intuitive to implement for some problems
- Choice between techniques depends on problem structure and implementation constraints
Space complexity optimization
- Reduces memory usage in dynamic programming algorithms
- Hirschberg's algorithm optimizes space complexity for sequence alignment
- Reduces space from to while maintaining time complexity
- Sliding window techniques for processing long sequences
- Memory-efficient backtracking for reconstructing optimal solutions
- Trade-offs between time and space complexity in algorithm design
Time complexity analysis
- Evaluates algorithm efficiency in terms of input size
- Common time complexities in bioinformatics dynamic programming:
- for pairwise sequence alignment (m, n are sequence lengths)
- for basic RNA structure prediction (n is sequence length)
- for some NP-hard problems (k is a constant, n is input size)
- Asymptotic analysis using Big O notation to describe worst-case scenarios
- Consideration of average-case and best-case complexities for real-world performance
Advanced dynamic programming concepts
- Extend basic dynamic programming techniques to address more complex bioinformatics problems
- Incorporate sophisticated models of biological processes and evolutionary relationships
- Enable more accurate and nuanced analysis of biological sequences and structures
Affine gap penalties
- Model biological insertions and deletions more realistically than linear gap penalties
- Use different costs for gap opening and gap extension
- Implemented in dynamic programming algorithms using additional matrices or states
- Improve alignment accuracy, especially for sequences with long insertions or deletions
- Applications include:
- Protein sequence alignment with large indels
- Genomic sequence alignment with intron modeling
Profile hidden Markov models
- Probabilistic models representing multiple sequence alignments
- Use dynamic programming for sequence alignment and probability calculation
- Three main states: match, insert, and delete
- Forward algorithm calculates probability of sequence given model
- Viterbi algorithm finds most probable path through model for a sequence
- Applications include:
- Protein family classification
- Sequence homology detection
- Multiple sequence alignment
Pair hidden Markov models
- Extend HMMs to model alignment between two sequences simultaneously
- Three emission states: match (aligned positions), insert X, insert Y
- Use dynamic programming for alignment and probability calculation
- Enable probabilistic sequence alignment and similarity assessment
- Applications include:
- Pairwise sequence alignment with uncertainty quantification
- Comparative gene finding
- Alignment of distantly related sequences
Limitations and alternatives
- Recognize constraints of dynamic programming in certain bioinformatics scenarios
- Explore complementary approaches to overcome limitations
- Combine multiple techniques for more robust and efficient solutions
Heuristic approaches
- Sacrifice guaranteed optimality for improved speed and scalability
- BLAST (Basic Local Alignment Search Tool) uses seed-and-extend heuristic
- Faster than Smith-Waterman but may miss some optimal alignments
- FASTA algorithm for rapid sequence database searching
- Approximate string matching algorithms for large-scale genomic comparisons
- Trade-offs between speed and sensitivity in heuristic methods
Probabilistic methods
- Incorporate uncertainty and variation in biological data analysis
- Bayesian inference for parameter estimation and model selection
- Markov Chain Monte Carlo (MCMC) methods for sampling complex distributions
- Maximum likelihood estimation for phylogenetic tree reconstruction
- Applications include:
- Protein structure prediction
- Gene regulatory network inference
- Population genetics analysis
Machine learning integration
- Combine dynamic programming with machine learning for enhanced performance
- Neural networks for improved scoring functions in sequence alignment
- Support Vector Machines (SVMs) for protein classification and function prediction
- Deep learning approaches for:
- Protein structure prediction (AlphaFold)
- Gene expression analysis
- Genomic variant calling
Software tools and libraries
- Provide implementations of dynamic programming algorithms for bioinformatics
- Enable researchers to apply complex algorithms without reimplementing from scratch
- Facilitate reproducibility and standardization in bioinformatics research
BLAST implementation
- NCBI BLAST+ suite implements various BLAST algorithms
- Uses dynamic programming for extending initial seed matches
- Optimized for speed and memory efficiency
- Supports multiple sequence databases and search types
- Applications include:
- Homology searching in genomic and protein databases
- Identification of conserved domains and motifs
- Metagenomics analysis
Bioconductor packages
- R-based software project for analysis of genomic data
- Biostrings package for string manipulation and pairwise alignment
- GenomicRanges for representing and manipulating genomic intervals
- DESeq2 for differential gene expression analysis
- Packages implement various dynamic programming algorithms for:
- Sequence alignment and comparison
- RNA-seq data analysis
- ChIP-seq peak calling
Custom algorithm development
- Biopython library provides tools for developing bioinformatics algorithms in Python
- SeqAn C++ library for sequence analysis algorithm implementation
- CUDA-based implementations for GPU-accelerated dynamic programming
- Considerations for custom development:
- Algorithm optimization for specific use cases
- Integration with existing bioinformatics pipelines
- Scalability for large-scale data processing
Future directions
- Explore emerging trends and potential advancements in dynamic programming for bioinformatics
- Address current limitations and challenges in the field
- Anticipate future needs and applications in biological data analysis
Parallel computing applications
- Harness multi-core CPUs and GPUs for accelerated dynamic programming
- Distributed computing frameworks (Hadoop, Spark) for large-scale genomic data processing
- Cloud computing platforms for on-demand scaling of bioinformatics workflows
- Parallel implementations of sequence alignment and structure prediction algorithms
- Challenges include:
- Algorithm redesign for effective parallelization
- Load balancing in heterogeneous computing environments
Deep learning enhancements
- Integrate deep learning models with dynamic programming algorithms
- Attention mechanisms for improved sequence alignment and comparison
- Transformers for context-aware sequence analysis and prediction
- Graph neural networks for protein structure and interaction prediction
- Potential applications:
- Enhanced protein function prediction
- Improved gene regulatory network inference
- More accurate protein-protein interaction prediction
Big data integration
- Develop algorithms to handle increasing volumes of biological data
- Integration of multi-omics data (genomics, transcriptomics, proteomics)
- Scalable dynamic programming approaches for population-scale genomics
- Machine learning techniques for feature extraction and dimensionality reduction
- Challenges include:
- Handling heterogeneous data types and formats
- Ensuring privacy and security of sensitive biological data
- Developing interpretable models for complex biological systems