Principal Component Analysis (PCA) is a powerful technique for reducing data complexity. It transforms high-dimensional datasets into lower-dimensional representations, preserving key information while simplifying analysis and visualization.
PCA finds patterns in data by identifying directions of maximum variance. This allows us to compress data, remove noise, and uncover hidden structures, making it a valuable tool in many fields like machine learning and data science.
PCA for Dimensionality Reduction
Fundamentals of Principal Component Analysis
- Principal Component Analysis (PCA) reduces high-dimensional data while preserving original variability
- Transforms original variables into uncorrelated principal components through linear combinations
- Identifies patterns in data highlighting similarities and differences
- Visualizes high-dimensional data by projecting onto lower-dimensional space (typically 2 or 3 dimensions)
- Applies to various fields (image processing, finance, bioinformatics, machine learning) for feature extraction and noise reduction
- Addresses curse of dimensionality by reducing features while retaining most information
- Assumes directions with largest variance contain most important aspects of data structure
Applications and Benefits of PCA
- Enables efficient data compression by representing data with fewer dimensions
- Improves performance of machine learning algorithms by reducing overfitting
- Facilitates data exploration and visualization of complex datasets
- Enhances signal processing by separating signal from noise
- Aids in feature selection by identifying most important variables
- Supports anomaly detection by revealing unusual patterns in reduced space
- Enables efficient data storage and transmission through dimensionality reduction
Applying PCA to Data Transformation
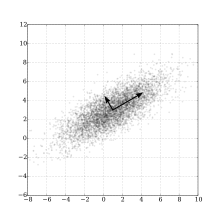
PCA Algorithm Steps
- Center data by subtracting mean of each variable from corresponding values
- Compute covariance matrix of centered data capturing relationships between variables
- Perform eigenvalue decomposition on covariance matrix obtaining eigenvectors and eigenvalues
- Sort eigenvectors in descending order of corresponding eigenvalues
- Construct transformation matrix by selecting top k eigenvectors for desired reduced dimensions
- Project original data onto new lower-dimensional space by multiplying with transformation matrix
Mathematical Foundations of PCA
- Eigenvectors represent directions of maximum variance in data
- Eigenvalues indicate amount of variance explained by each eigenvector
- PCA maximizes variance of projected data subject to orthogonality constraint
- Covariance matrix C of centered data X calculated as
- Eigenvalue problem solved through equation
- Transformation matrix W formed by concatenating selected eigenvectors
- Reduced data Y obtained by matrix multiplication
Interpreting Principal Components
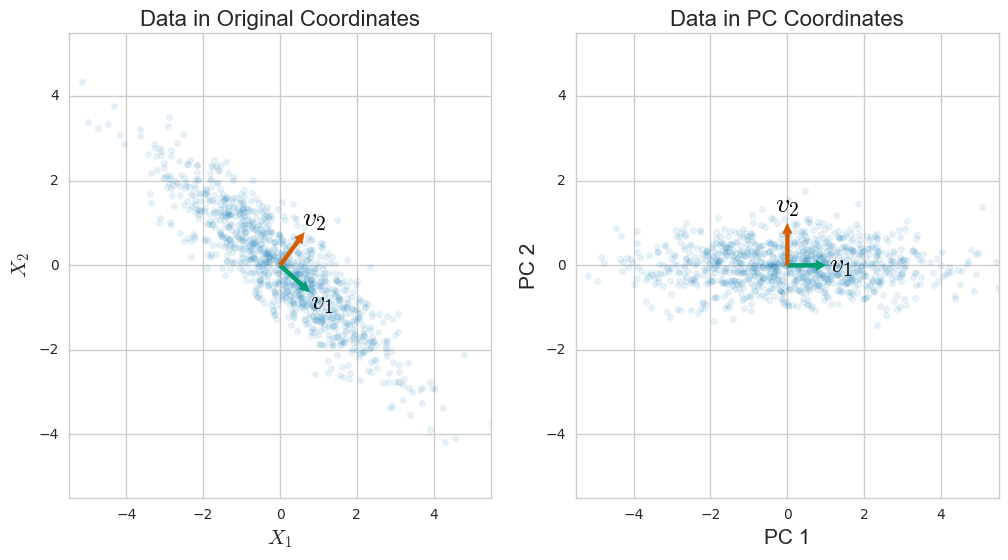
Understanding Principal Component Significance
- Each principal component represents direction of maximum data variance orthogonal to previous components
- First principal component accounts for largest amount of variance with subsequent components explaining progressively less
- Loadings of original variables on principal component indicate contribution to component
- Proportion of variance explained by each component calculated by dividing eigenvalue by sum of all eigenvalues
- Scree plot graphs eigenvalues against component number to visually determine significant components
- Cumulative proportion of variance explained determines components needed to retain desired percentage of original information
- Biplots visualize observations and original variables in reduced space showing relationships and contributions to principal components
Analyzing Principal Component Composition
- Examine magnitude and sign of loadings to interpret component meaning
- Large positive loadings indicate strong positive correlation with component
- Large negative loadings indicate strong negative correlation with component
- Small loadings suggest minimal influence on component
- Compare loadings across components to identify variables contributing to multiple dimensions
- Analyze patterns in loadings to uncover underlying structures or latent factors in data
- Consider domain knowledge when interpreting component meanings and significance
Evaluating PCA Effectiveness
Quantitative Measures of PCA Performance
- Total variance explained by selected principal components measures information preservation
- Reconstruction error quantifies information loss due to dimensionality reduction
- Kaiser criterion retains components with eigenvalues greater than 1
- Cross-validation assesses generalizability of PCA model to unseen data
- Compare PCA results with alternative dimensionality reduction techniques (t-SNE, UMAP)
- Measure computational efficiency and scalability for large datasets
- Evaluate stability of principal components through bootstrapping or perturbation analysis
Considerations for PCA Application
- Interpretability of principal components crucial for determining effectiveness in problem domain
- Sensitivity to outliers may affect results in datasets with extreme values
- Assumption of linearity limits effectiveness for strongly non-linear relationships
- Scaling of input variables impacts PCA results consider standardization or normalization
- Balance between dimensionality reduction and information retention based on specific application needs
- Assess impact of PCA on downstream tasks (classification, clustering) to gauge overall effectiveness
- Consider domain-specific metrics or visualizations to evaluate PCA performance in context